The human body is constantly responding to oxidative stressors. If these stressors, such as pollution, infection, microorganisms, smoking, and even our diet, aren’t controlled cellular damage can occur leading to cancer, neurodegenerative diseases, and aging. As this battle rages within our bodies, cells reprogram translation, shutting down the production of major housekeeping proteins. This helps conserve energy and resources so cells can produce anti-oxidant proteins to combat the stressor’s harmful effects. It’s a never-ending cycle the body repeats to regain homeostasis.
It was previously thought that in order to cease stress-induced protein production, only one mechanism of inhibition at the initiation stage was sufficient. However, roughly 15 years ago, scientists realized there might be something more beyond initiation—they just didn’t know what it was.
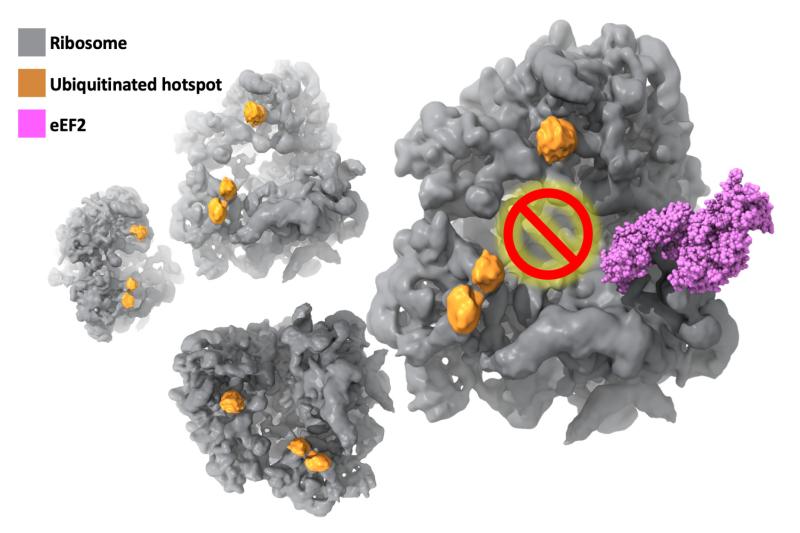
Now, findings from an international cooperation between Duke University and several US and European Institutes* has provided the first evidence of a new mechanism involving K63 ubiquitination of ribosomes that represses protein production during the elongation stage, and is essential in combatting oxidative stress.
Using budding yeast as a model, Gustavo Silva, PhD, of Duke Biology worked closely with Duke Biochemistry’s Alberto Bartesaghi, PhD, leveraging his team’s cryo-EM image and computational expertise to solve the molecular structure of ribosomes under stress at an unprecedented level of detail. Cryo-EM allowed purification of the ribosomes computationally rather than biochemically and allowed researchers to take snapshots at multiple states of translation—piecing them together to reveal how cells react under stress.
The fact that Silva and Bartesaghi were at the same university helped accelerate the process. Bartesaghi’s postdoc and first author of the manuscript, Ye Zhou, analyzed the images collected at the Shared Materials Instrumentation Facility using high-performance computing resources provided by Duke Research Computing and met weekly with the two researchers to interpret the final 3D structures.
t took the combination of multiple labs and cryo-EM to find the missing link. The next step is to understand the function of ubiquitinated ribosome subpopulations which could establish ubiquitin as a master regulator of protein production. Silva’s lab is now using human cells as a model, which in combination with techniques for visualizing proteins at atomic resolution developed in Bartesaghi’s lab, will bring us one step closer to developing drugs that regulate protein production, increase cell health, and can be used to prevent cancer, neurodegenerative diseases, or just slow down the aging process.
Read more about this research in the latest issue of PNAS
*This research was done in collaboration with the National Institute of Environmental Health Sciences (NIH); Center for Genomics and Systems Biology (NYU); Martin Luther University Halle-Wittenberg; Halle/Saale, Germany, Max-Planck Institute of Biochemistry; European Molecular Biology; Bijvoet Center for Biomolecular Research.